DIMENSION REDUCTION AND SHRINKAGE
1. VARIABLE SELECTION
> attach(Auto)
> library(leaps)
> reg.fit
= regsubsets(
mpg ~ cylinders + displacement + horsepower + weight + acceleration + year,
Auto )
> summary(reg.fit)
Selection Algorithm: exhaustive
cylinders
displacement horsepower weight acceleration year
1 (
1 ) " " "
" " " "*" " " " "
2 (
1 ) " " "
" " " "*" " " "*"
3 (
1 ) " " "
" " " "*" "*" "*"
4 (
1 ) " "
"*" "
" "*" "*" "*"
5 (
1 ) "*"
"*" "
" "*" "*" "*"
6 (
1 ) "*"
"*"
"*"
"*" "*" "*"
# This command finds
the best model for each p = number of independent variables. The best model is
determined by the lowest RSS.
# Next, choose the best
p according to some criteria:
> summary(reg.fit)$adjr2 # Adjusted R2
[1] 0.6918423 0.8071941 0.8071393 0.8067872 0.8067841 0.8062826
> summary(reg.fit)$cp # Mallows Cp
[1] 232.396144 1.169751 2.284200 3.992019 5.000800 7.000000
> summary(reg.fit)$bic # BIC = Bayesian information criterion
[1] -450.5016 -629.3564 -624.2828 -618.6081 -613.6448 -607.6743
# Recall that plain R2 is not
a fair measure of performance. It always increases with p:
> summary(reg.fit)$rsq
[1] 0.6926304 0.8081803 0.8086190 0.8087638 0.8092549 0.8092553
# For
stepwise or backward elimination variable selection, use method=”forward” or
method=”backward”.
> library(MASS)
> reg =
regsubsets( medv ~ .,
data=Boston, method =
"backward" )
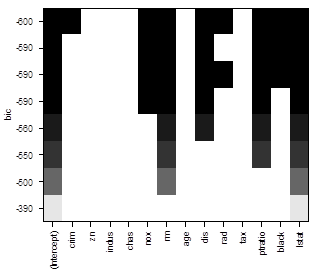
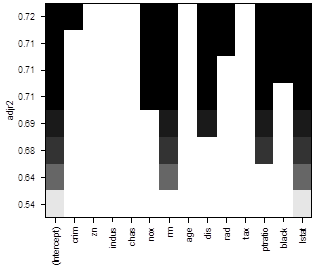
# There is a nice way
to visualize results, ranking models by the chosen “scale”. Black color means
the variable is included into the model, white means it is excluded.
> plot(reg)
> plot(reg,
scale = "adjr2" )
# To see more models, use option “nbest”, which is the number of models of each size p to be
compared.
> reg = regsubsets( medv ~ ., data=Boston, method = "backward", nbest=4 )
> plot(reg, scale = "adjr2" )
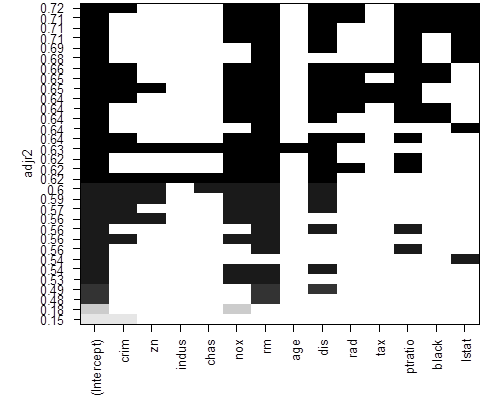
# We can also choose
the best model by means of a stepwise procedure, starting with one model and
ending with another.
> null
= lm( medv ~ 1, data=Boston )
> full
= lm( medv ~ ., data=Boston )
> step( null, scope=list(lower=null,
upper=full), direction="forward" )
Start: AIC=2246.51
medv ~ 1
Df Sum of Sq RSS AIC
+ lstat 1 23243.9 19472 1851.0 # Compare
contributions of
+ rm 1 20654.4 22062 1914.2 # remaining
independent variables
+ ptratio 1
11014.3 31702 2097.6
+ indus 1 9995.2 32721 2113.6
+ tax 1
9377.3 33339 2123.1
+ nox 1 7800.1 34916 2146.5
+ crim 1 6440.8 36276 2165.8
+ rad 1
6221.1 36495 2168.9
+ age 1
6069.8 36647 2171.0
+ zn 1 5549.7 37167 2178.1
+ black 1
4749.9 37966 2188.9
+ dis 1
2668.2 40048 2215.9
+ chas 1 1312.1 41404 2232.7
<none> 42716 2246.5
Step: AIC=1851.01
medv ~ lstat
Df Sum of Sq RSS AIC
+ rm 1 4033.1 15439 1735.6
+ ptratio 1
2670.1 16802 1778.4
+ chas 1 786.3 18686 1832.2
+ dis 1
772.4 18700 1832.5
+ age 1
304.3 19168 1845.0
+ tax 1
274.4 19198 1845.8
+ black 1
198.3 19274 1847.8
+ zn 1
160.3 19312 1848.8
+ crim 1 146.9 19325 1849.2
+ indus 1 98.7 19374 1850.4
<none> 19472 1851.0
+ rad 1
25.1 19447 1852.4
+ nox 1 4.8 19468 1852.9
... < truncated > ...
Step: AIC=1585.76
medv ~ lstat + rm + ptratio + dis + nox + chas + black + zn +
crim
+ rad + tax
Df Sum of Sq RSS AIC
<none> 11081 1585.8
+ indus 1
2.51754 11079 1587.7
+ age 1
0.06271 11081 1587.8
Call:
lm(formula = medv ~ lstat + rm + ptratio + dis + nox + chas +
black + zn + crim + rad + tax, data =
Boston)
Coefficients:
(Intercept) lstat rm ptratio dis nox
36.341145
-0.522553 3.801579 -0.946525
-1.492711 -17.376023
chas black zn crim rad tax
2.718716
0.009291 0.045845 -0.108413 0.299608
-0.011778
# The final model contains variables
lstat,
rm, ptratio, dis, nox, chas, black, zn, crim, rad, and tax.
2. RIDGE REGRESSION
# Dataset “Boston” has some strong correlations,
resulting in multicollinearity.
> cor(Boston)
# Apply ridge regression
> lm.ridge( medv~., data=Boston, lambda=0.5
)
crim
zn
indus
chas
36.270091954 -0.107524388 0.046092635
0.018815242 2.693518778
nox rm age dis rad
-17.646013524 3.816019080
0.000578687 -1.469191794 0.301928336
tax ptratio black lstat
-0.012134702 -0.950831885 0.009309388
-0.523845421
# We can see how the slopes
change with penalty lambda. These are estimated slopes for different lambda.
When lambda=0, we get LSE. Large lambda forces them toward 0.
> rr =
lm.ridge(medv ~ .,
data=train, lambda=seq(0,1000,1) )
> rr
> plot(rr)
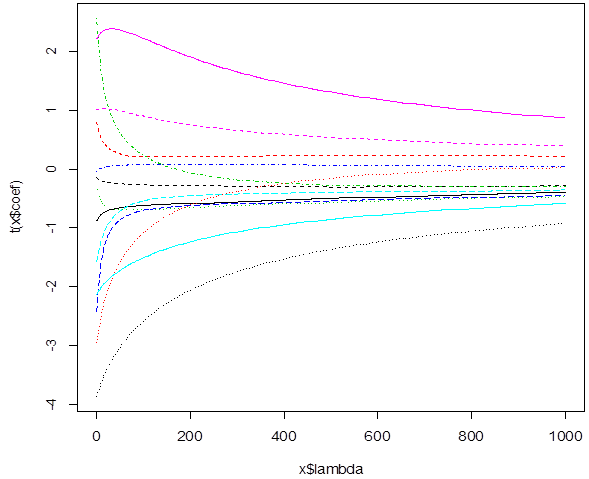
# To choose a good lambda, fit
ridge regression with various lambda and compare prediction performance.
>
select(rr)
modified HKB estimator is 4.321483
modified L-W estimator is 3.682107
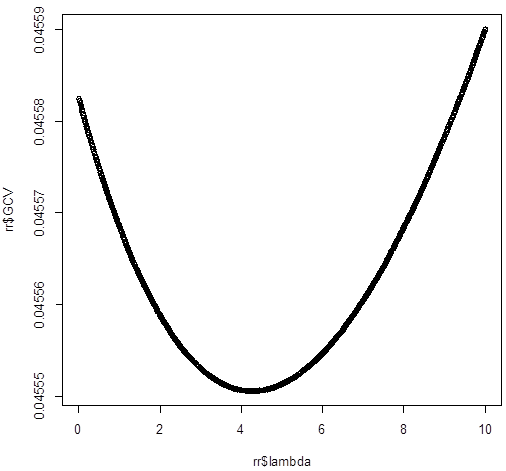
smallest value of GCV at 5
# So, the best lambda is
around 5. We’ll look closer at that range.
>
rr = lm.ridge(
medv~., data=Boston, lambda=seq(0,10,0.01)
)
>
select(rr)
modified HKB estimator is 4.594163
modified L-W estimator is 3.961575
smallest value of GCV at 4.26
>
plot(rr$lambda,rr$GCV)